Some Boring Analysis on TLGS Index (and messing with CERN's ROOT analysis framework)
Lupa[1] provides constant basic statistics about the Gemini network. Stats like how many servers are online, number of virtual hosts, TLS distribution, etc.. It's a really cool project. It got me asking.. what kinds of information can I extract from TLGS' index? Furthermore, I want to provide some more information for Gemini users. Helping us to understand the network we love.
It also helps me to build a better search engine. There's no hero here.
First things first, I decided to do analysis using CERN's ROOT data analysis framework[2] instead of the more traditional Python + pandas. Foremost, I wanted to revist the framework because it helped me back in university doing some wicked things. Like sorting through TBs of CSV and run analysis on that. Without batching or ever running out of memory. Secondly, I intend (hopefully) to turn this project into an automated script and generates PDF as output. ROOT is sutable for this purpose as it can save plots as LaTeX.
ROOT is far from perfect. It's a bit rigid at times and the legacy interface is unpleasant. While the modern interface is not as featureful. I'll see what I can do.
Getting data into ROOT
First, I need to get the index from PostgreSQL into SQLite. ROOT's RDataFrame should support PostgreSQL in the future. But for now SQLite is the best option we have. The table format can be directly extracted from the source code of tlgs_ctl and use with minimal modification. Dumping the actual data is more complicated. What's generated by pg_dump is quite close to what SQLite can handle. But some changes are needed. I ended creating my own command to remove unsupported commands. And generates "dump.sql" as output.
pg_dump --data-only --inserts tlgs | grep -v -e "\\(.setval\\|^SET\\)" |\
sed "s/public\\.//g" | awk 'BEGIN {print "BEGIN;"} END{print "END;"} {print}' \
> dump.sql
With the data ready. I can apply the tables in SQLite and import the data.
CREATE TABLE IF NOT EXISTS pages (
url text NOT NULL,
domain_name text NOT NULL,
port integer NOT NULL,
content_type text,
charset text,
lang text,
title text,
content_body text,
size integer DEFAULT 0 NOT NULL,
last_indexed_at timestamp without time zone,
last_crawled_at timestamp without time zone,
last_crawl_success_at timestamp without time zone,
last_status integer,
last_meta text,
first_seen_at timestamp without time zone NOT NULL,
search_vector tsvector,
cross_site_links json,
internal_links json,
title_vector tsvector,
last_queued_at timestamp without time zone,
indexed_content_hash text NOT NULL default '',
raw_content_hash text NOT NULL default '',
PRIMARY KEY (url)
);
CREATE TABLE IF NOT EXISTS links (
url text NOT NULL,
host text NOT NULL,
port integer NOT NULL,
to_url text NOT NULL,
to_host text NOT NULL,
to_port integer NOT NULL,
is_cross_site boolean NOT NULL
);
CREATE TABLE IF NOT EXISTS robot_policies (
host text NOT NULL,
port integer NOT NULL,
disallowed text NOT NULL
);
CREATE TABLE IF NOT EXISTS robot_policies_status (
host text NOT NULL,
port integer NOT NULL,
last_crawled_at timestamp without time zone NOT NULL,
have_policy boolean NOT NULL,
PRIMARY KEY (host, port)
);
.read dump.sql
The result is not perfect. Some syntax errors here and there. But most of the data arrived intact.
The boring analysis
Now we can start messing with ROOT. Nowadays, analists are commonly asked to tell a story. I'll try that. As a search engine developer, I want to how large an average page is. Due to PostgreSQL's highlighting limitation. Currently highlighting is limited to the first 5K characters. I surely hope pages are not too large. In code, I create a dataframe with all text/gemini pages. Then create a histogram of the page sizes less then 20K. Draw it. Then print how many pages are not included in the histogram.
root [0] rdf = ROOT::RDF::MakeSqliteDataFrame("./sqlite.db"
, "SELECT size FROM pages WHERE content_type = \"text/gemini\"");
root [1] f1 = rdf.Filter("size != 0 && size < 20*1000")
(ROOT::RDF::RInterface<ROOT::Detail::RDF::RJittedFilter, void> &) @0xffff950ef088
root [2] h1 = f1.Histo1D("size")
(ROOT::RDF::RResultPtr<TH1D> &) @0xffff950ef0f8
root [3] h1->SetTitle("Distribution of text/gemini page size")
root [4] h1->Draw()
Info in <TCanvas::MakeDefCanvas>: created default TCanvas with name c1
root [5] c1->SaveAs("c1.png")
Info in <TCanvas::Print>: png file c1.png has been created
root [6] root [6] *rdf.Count() - *f1.Count()
(unsigned long long) 1550
In all of TLGS' index. There's only 1550 pages larger than 20K. And the distribution looks like the following. Nice! Looks like indeed most pages are quite small. And 5K is a good cutoff.
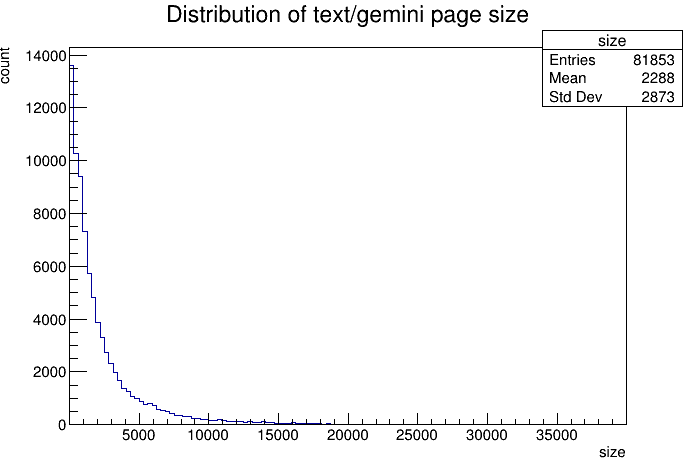
One question one may ask. Larger files tends to contain more information. Whcih makes it easier to be searched. Could we compensate for this? Yes! The data looks log-normal. Seems the Landau distribution will work. Assuming the number of tokens is corrilated to the size of the page. We can fit a curve to our distribution then multiply that curve against a linear function to get the expected result distribution (pivoting off the mean). Let's try it.
root [7] h1->Fit("landau")
FCN=617.185 FROM MIGRAD STATUS=CONVERGED 177 CALLS 178 TOTAL
EDM=2.35903e-08 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 Constant 7.21594e+04 8.50337e+02 3.07065e+00 -3.76367e-07
2 MPV 9.61922e+01 1.82522e+01 7.72726e-02 -2.20279e-05
3 Sigma 4.87176e+02 2.47542e+00 4.86057e-06 -3.43981e-01
(TFitResultPtr) <nullptr TFitResult>
root [9] sizeFunc = TF1("sizeFunc", "x/2288.0", 0, 20*1000)
(TF1 &) Name: sizeFunc Title: x/2288.0
root [10] fitFunc = *(h1->GetFunction("landau"))
(TF1 &) Name: landau Title: landau
root [11] correctedFunc = TF1("correctedFunc", "fitFunc(x)*sizeFunc(x)", 0, 20*1000)
(TF1 &) Name: landau Title: correctedFunc
root [12] correctedFunc.SetLineColor(3)
root [13] correctedFunc.Draw("SAME");
root [14] c1->SaveAs("corrected.png");
Info in <TCanvas::Print>: png file corrected.png has been created
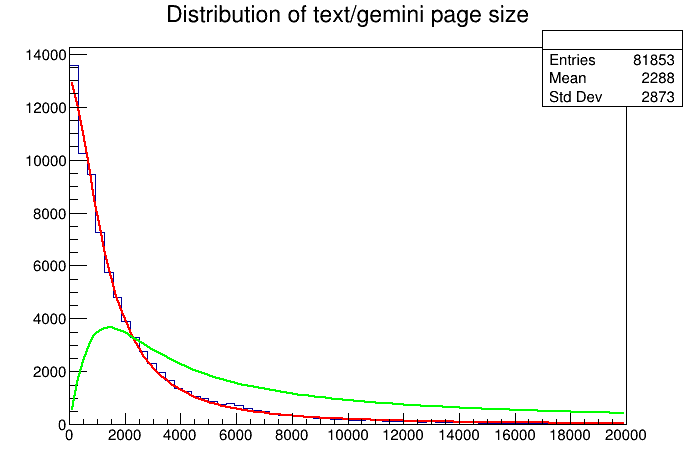
I'm too lazy to add a legend to the plot. The red curve is the fitted distribution. And the green one is the "corrected" distribution. Now limiting the highlighting to the first 5K characters seems to not be a good idea after all. Seems to be much space to the right. Hmm... But how much? Well, we can ask ROOT to inspect the curve.
root [15] i1 = correctedFunc.Integral(0, 5000)
(double) 13513212.
root [16] i2 = correctedFunc.Integral(0, 20*1000)
(double) 26138316.
root [16] i1/i2
(double) 0.51698864
Given my assumptions are correct. Only 51% of the pages will have "correct" highlighting. How about 10K? correctedFunc.Integral(0, 10000)/i2 prints 0.76409630, or 76%. That's a good indication that I shoud up the limit a bit.
Now we know the distribution of sizes. I wish to know the distribution of file size vs number of lines (remember Gemtext is a line based language).
root [0] rdf = ROOT::RDF::MakeSqliteDataFrame("./sqlite.db"
, "SELECT size, content_body FROM pages WHERE content_type = \"text/gemini\"");
root [1] f1 = rdf.Filter("size != 0 && size < 20*1000")
(ROOT::RDF::RInterface<ROOT::Detail::RDF::RJittedFilter, void> &) @0xffff950ef088
root [2] countOccur = [](const std::string &s) { return std::count(s.begin(), s.end(), '\n'); }
((lambda) &) @0xffffaa9e8018
root [3] f2 = f1.Define("lines", "countOccur(content_body)")
(ROOT::RDF::RInterface<ROOT::Detail::RDF::RJittedFilter, ROOT::RDF::RInterface<ROOT::Detail::RDF::RJittedFilter, void>::DS_t>) @0xaaaae2ef7b40
root [4] g1 = f2.Graph("size", "lines")
(ROOT::RDF::RResultPtr<TGraph> &) @0xffffae08d170
root [5] g1->Draw("ap")
Info in <TCanvas::MakeDefCanvas>: created default TCanvas with name c1
root [6] c1->SaveAs("scatter.png")
Info in <TCanvas::Print>: png file scatter.png has been created
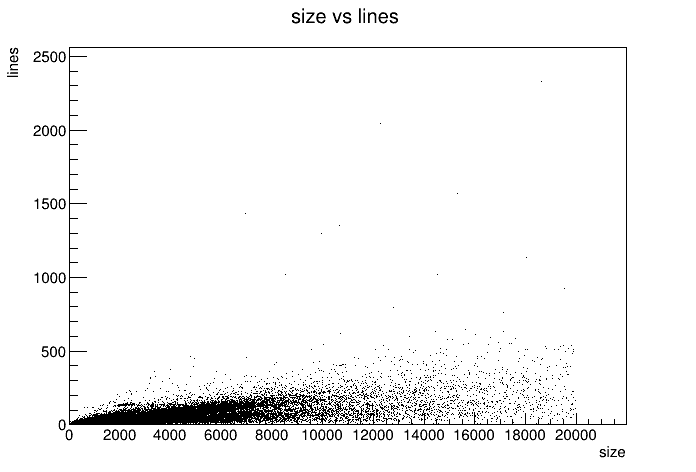
Wow that's.. underwhelming. Sure there's a trend of more equal larger file size. But there's no intresting trend. Unless you count files eeming to be 20K in size but only have a few lines. I guess no result is also a result. I guess that just means there's very different ways how people write on Gemini. Maybe this points to machine generated content? Some uber long URL? Maybe worth future investigation. But not today. Nevertheless, I want a histogram of the distribution.
root [7] h2 = f2.Histo2D({"h1", "size vs lines", 64, 0, 128, 128, 20000, 2500}, "size", "lines")
(ROOT::RDF::RResultPtr<TH2D> &) @0xffffae08d220
root [8] h2->Draw("LEGO1")
root [9] c1->SaveAs("h2d.png")
Info in <TCanvas::Print>: png file h2d.png has been created
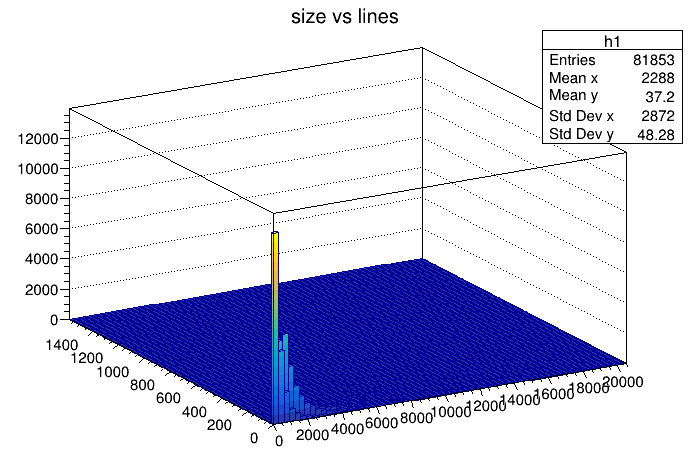
I should have gone with a histogram in the first place. It more clearly shows the distribution - there's a lot of small files and some medium sized files. Big ones are rare. I should have expected this. The 1st histogram already show small file sizes.
Now finished with a single page. I want to look at the connection between pages. To be specific, what's the distribution of outward backlinks? The code for this will be a bit more complicated. And uses more memory. But should be fine.
root [0] rdf = ROOT::RDF::MakeSqliteDataFrame("./sqlite.db", "SELECT pages.url AS url FROM "
"pages JOIN links ON pages.url=links.to_url WHERE links.is_cross_site = TRUE "
"AND content_type = \"text/gemini\"")
(ROOT::RDataFrame &) A data frame associated to the data source "generic data source"
root [1] unordered_map<string, size_t> inlink_count
(std::unordered_map<std::string, size_t> &) {}
root [2] rdf.Foreach([](const std::string& url) {inlink_count[url]++;}, {"url"})
root [3] inlink_count.size()
(unsigned long) 12687
root [4] vector<float> counts
(std::vector<float> &) {}
root [5] for(auto& [_, v] : inlink_count) counts.push_back(v);
root [6] h1 = TH1F("h1", "histogram of inlinks", 128, *min_element(counts.begin(), counts.end()), *max_element(counts.begin(), counts.end()))
(TH1F &) Name: h1 Title: histogram of inlinks NbinsX: 128
root [7] for(auto v : counts) h1.Fill(v)
root [8] h1.Draw()
Info in <TCanvas::MakeDefCanvas>: created default TCanvas with name c1
root [9] c1->SaveAs("inlink_hist.png")
Info in <TCanvas::Print>: png file inlink_hist.png has been created
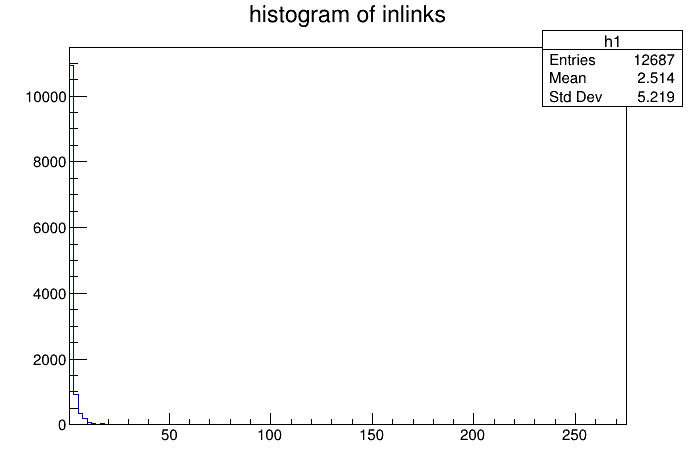
Unsuprisingly, most pages that does get a backlink have one (since pages with 0 backlinks will not register in our SQL query). I'm super surprised that there isn't more pages with more backlinks. I assum sites like geminispace.info, medusae.space, etc.. would have a lot of backlinks. I think the bin does not overlap thus not showing up in a histogram. Suprisingly, there isn't that much cross-capsule backlinks. 12687 is the total on the entire index. That's super low considering the amount of work people put into their content and aggregrators generating backlinks.
I noticed I chould have generated the backlink count per-site directly in SQL. Welp.
Finally, I want to know how compressible Gemini content is and by extention, how large is the content indexed. TLGS removes ASCIi arts and other unneeded text before indexing. Thus the indexed size is different than the original size. But how do we check the compression ratio? Importing zlib and using it would be to complicated. Instead, ROOT has it's own data archive format with the .root extension. It applies LZ4 compression to all stored data. Which we could use as a upper bound for the compression ratio (Since it has to store other metadata).
root [0] rdf = ROOT::RDF::MakeSqliteDataFrame("./sqlite.db",
"SELECT size, content_body FROM pages WHERE content_type = \"text/gemini\"");
(ROOT::RDataFrame &) A data frame associated to the data source "generic data source"
root [7] original_size = *rdf.Sum("size")
(double) 5.3742992e+08
root [8] indexed_size = *rdf.Define("indexed_size"
, [](const string& s) {return s.size();}, {"content_body"}).Sum("indexed_size")
(double) 2.3638077e+08
root [10] rdf.Snapshot("content", "content.root", {"content_body"})
(ROOT::RDF::RResultPtr<ROOT::RDF::RInterface<ROOT::RDF::RInterface<ROOT::Detail::RDF::RLoopManager,
void>::RLoopManager> >) @0xaaaaeabc1a90
root [11] .! ls -lh content.root
-rw-r--r-- 1 marty marty 97M Jul 29 12:51 content.root
I'm (again) surprised how small Gemini is. Only 537MB of text is in my index. Moreover, only 236MB after removing ASCII art. More than a 50% reduction. Wow. The ~41% compression ratio upper bound looks about right. The typical ratio we'd expect to pure text.
Conclusion
Few things learned from this project.
- I might want to increase the limit of text highlighting to 10K.
- Some large files have very few lines
- There're a lot of small files. But not really searchable by search engine
- Gemini is short on backlinks
- The smol internet is smol
That's all. Martin out